Table of Contents |
Apart from regulatory processes that you will learn about in other lessons, the first step in gene expression is the copying of information in DNA into RNA. This process is called transcription. The reverse case of copying information in RNA into DNA, which is used by some viruses and laboratory techniques, is called reverse transcription, and the DNA product is called complementary DNA (cDNA). The product of transcription is called a transcript.
When a region of DNA is unwound to allow transcription to take place, the transcript is formed through complementary base pairing with one strand of DNA. Complementary base pairing during transcription is similar to the process used during DNA replication except that each DNA nucleotide is paired with an RNA nucleotide (a ribonucleotide) and adenine is paired with uracil instead of thymine.
The transcribed strand of DNA is called the template or sense strand. The other strand of DNA, which is not transcribed, is called the coding or antisense strand.
In bacteria, a single type of RNA polymerase (DNA-dependent RNA polymerase) is used to pair complementary nucleotides to a template strand of DNA to form a complementary RNA transcript. Like DNA polymerase, RNA polymerase adds nucleotides one by one to the 3′-OH group of the growing nucleotide chain. Each nucleotide is joined to the preceding nucleotide by a covalent phosphodiester bond formed by dehydration synthesis.
However, an important difference between RNA polymerase and DNA polymerase is that RNA polymerase does not require a free 3′-OH to add a nucleotide. This means that, unlike DNA polymerase, it does not require a primer to begin forming a strand of RNA.
In Escherichia coli bacteria, a single RNA polymerase enzyme consists of six polypeptide subunits. Five of these subunits form the polymerase core enzyme responsible for adding RNA nucleotides to the growing strand. The sixth subunit, called the sigma (σ) factor or sigma subunit, enables RNA polymerase to bind to a specific sequence of DNA called a promoter. Different sigma factors enable the enzyme to bind to different promoters and therefore influence which genes are expressed.
IN CONTEXT
There are three steps of transcription in prokaryotes. During initiation of transcription, RNA polymerase and the other transcription machinery bind to a promoter. During elongation in transcription, the sigma subunit falls off and RNA polymerase adds nucleotides to produce a growing strand. Finally, termination of transcription concludes the process, and the new transcript is released.
Initiation of transcription begins when RNA polymerase binds to a promoter (a sequence of DNA). The specific nucleotide pair in the DNA double helix that corresponds to the site from which the first 5′ RNA nucleotide is transcribed is called the initiation site. Nucleotides preceding that site are designated “upstream”, whereas nucleotides following the initiation site are called “downstream” nucleotides. After RNA polymerase binds, it moves downstream as it adds nucleotides.
Most promoters are located just upstream of the genes they regulate, and these positions are described by a value representing the number of nucleotides below the initiation site (designated as position +1). Positions of downstream elements are described by a positive value representing the number of nucleotides from the initiation site.
Although promoter sequences vary, a few elements are highly similar across many organisms (conserved), and these are called consensus sequences. These sequences often have very important functions, meaning that mutations in them are unlikely to persist because they are so detrimental.
There are two promoter consensus sequences in eukaryotes at the −10 and −35 positions. The −10 consensus sequence is called the TATA box and its general sequence is TATAAT. Both the -10 and −35 consensus sequences are recognized and bound by sigma.
The image below shows elongation in transcription. This stage begins as soon as the sigma subunit dissociates from the core enzyme of the RNA polymerase performing transcription. The DNA must be separated to form a transcription bubble to allow a complementary strand to be formed based on the template strand. The new strand of RNA is synthesized beginning at the 5′ end and proceeding in a 5′ to 3′ direction. The double helix is unwound ahead of the core enzyme to allow complementary base pairs to be added. As transcription proceeds, the transcript separates from the template strand and the double helix reforms behind the RNA polymerase.
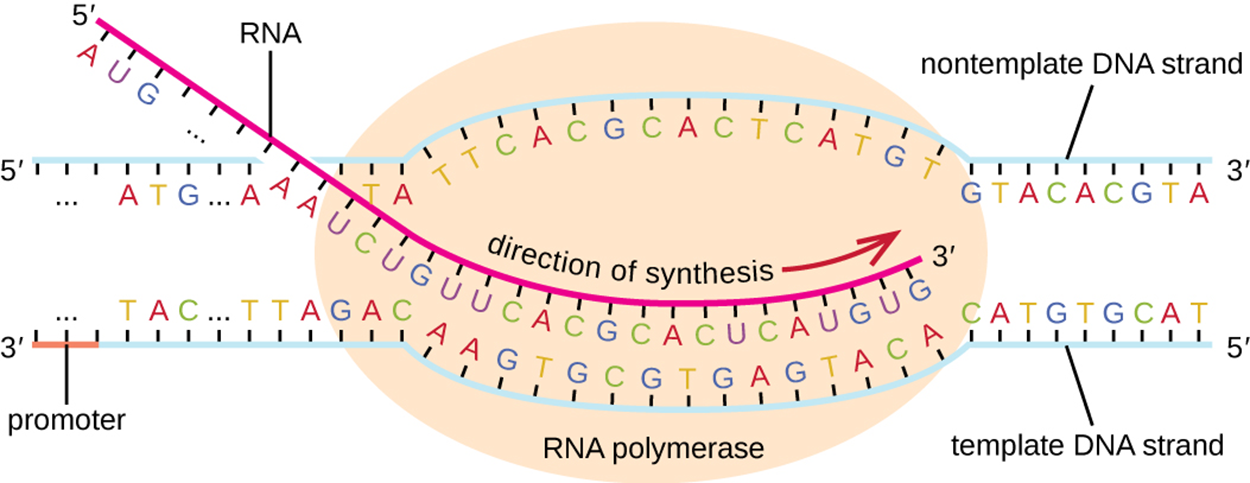
Transcription ends with termination. During this step, RNA polymerase dissociates from the DNA template and the RNA transcript is released. The DNA template includes repeated nucleotide sequences that act as termination signals, causing RNA polymerase to stall and release the DNA template. This frees the newly formed transcript.
There are important differences in transcription by domain and we will focus primarily on a comparison of bacteria versus eukaryotes. For example, bacterial RNA polymerase differs from each of the three RNA polymerases used by eukaryotes (RNA polymerases I, II, and III). Archaea have a single RNA polymerase, but it is more similar to eukaryotic RNA polymerase II than to bacterial RNA polymerase. Additionally, eukaryotic mRNAs are usually monocistronic (i.e., they encode one polypeptide), whereas both bacteria and archaea tend to have polycistronic mRNAs (i.e., they encode multiple polypeptides).
In prokaryotes, transcription takes place in the cytoplasm and the newly produced transcripts can be used immediately to produce proteins. In eukaryotes, transcription takes place in the nucleus and protein-encoding transcripts must be transported from the nucleus to the cytoplasm before they can be used to make proteins.
In eukaryotes, protein-encoding mRNA is produced by the processing of primary transcripts (the RNA molecules directly transcribed by RNA polymerase). Processing can make the transcripts more stable and eukaryotic mRNAs can have much longer lifespans than bacterial mRNAs.
The table below summarizes major differences in transcription between bacteria and eukaryotes.
| Comparison of Transcription in Bacteria and Eukaryotes | ||
|---|---|---|
| Property | Bacteria | Eukaryotes |
| Number of polypeptides encoded per mRNA | Monocistronic or polycistronic | Exclusively monocistronic |
| Strand elongation | Single RNA polymerase | RNA polymerases I, II, or III |
| Addition of 5′ cap | No | Yes |
| Addition of 3′ poly-A tail | Yes | Yes |
| Splicing of pre-mRNA | No | Yes |
Note that the video states that prokaryotes are polycistronic and eukaryotes are monosaccharides because this is usually the case, but, as mentioned in the table above, there are exceptions.
Source: THIS CONTENT HAS BEEN ADAPTED FROM OPENSTAX’s “MICROBIOLOGY”. ACCESS FOR FREE AT openstax.org/details/books/microbiology
REFERENCES
Hajnsdorf, E., & Kaberdin, V. R. (2018). RNA polyadenylation and its consequences in prokaryotes. Philosophical transactions of the Royal Society of London. Series B, Biological sciences, 373(1762), 20180166.
doi.org/10.1098/rstb.2018.0166