In this lesson, you will learn about ways in which biochemistry can be used to identify microbes in the laboratory, focusing on bacteria. Many bacteria look similar or identical under the microscope, so other approaches are needed to accurately identify them. Identification can be critical. For example, it may be necessary to determine what pathogen is causing a disease to select the best antibiotic to treat an infection. In some cases, biochemical approaches can be highly effective to identify microbes from specimens. Specifically, this lesson will cover the following:
1. Introduction to the Use of Biochemistry for Identification
It is often important to identify microbes for study. For example, it may be necessary to determine what pathogen is causing an illness to trace its spread or to choose a treatment approach.
Some microbes can be easily identified by their appearance, but others look similar or identical under the microscope. For example, it is often difficult to identify bacteria under the microscope based on shape and appearance. Staining may help, but generally is used to classify bacteria into groups (such as gram positive versus gram negative) rather than for identification. In many cases, it takes multiple steps to identify a bacterium. These steps may include genotypic identification and biochemical characteristics.
Varied approaches are available that use biochemical characteristics to aid in identification. Some of these approaches involve determining whether a particular substance is present. For example, analysis of the contents of cytoplasmic granules can be useful in identifying certain bacteria. The presence or absence of the carbon- and energy-storage compound poly-β-hydroxybutyrate (PHB) and the presence or absence of fluorescent pigments can be helpful in identifying some species of the genus Pseudomonas.
Other approaches examine biochemical reactions used by different microbes. These include carbon utilization and other metabolic tests. These may be carried out individually, testing one reaction at a time, or using a system that allows panels of biochemical reactions to be carried out simultaneously and analyzed using software. Identifications can be performed manually or using automation.
-
EXAMPLE
There are many different types of biochemical and metabolic tests that can be used. Prior to the availability of modern technologies, these were a mainstay of microbial identification.
One common test examines whether a microbe produced the enzyme catalase. This enzyme catalyzes the conversion of hydrogen peroxide into water and oxygen. It is an important enzyme for microbes that live in an oxygen-rich environment.

The test is performed by adding hydrogen peroxide to a sample, then watching. If catalase is present, the newly produced oxygen forms clearly visible bubbles, indicating Gram-positive
Staphylococcus. If the catalase is not present, then no bubbling occurs, indicating Gram-positive
Streptococcus (e.g., Saginur et al., 1982).
2. MALDI-TOF Analysis
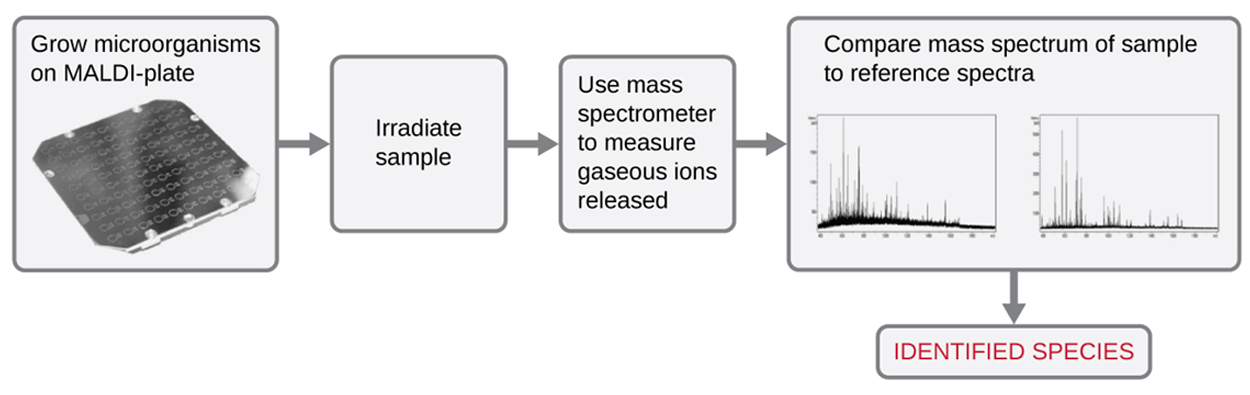
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) is an automated approach to compare the mass spectrum (the mass-to-charge ratios of ions in a sample) of a specimen to a database of known mass spectra. The procedure used is described in the steps and shown in the figure below.
-
1. The microbe is mixed with a specialized matrix reagent on a disposable MALDI plate.
2. The sample/reagent is irradiated with a high-intensity pulsed ultraviolet laser, resulting in the ejection of gaseous ions generated from the chemical constituents of the microbe.
3. The gaseous ions are collected and accelerated through the mass spectrometer, with ions traveling at a velocity determined by their mass-to-charge ratio (
m/z) and therefore reaching the detector at different times.
4. Detector signal is plotted against
m/z to yield a mass spectrum of the organism that is uniquely related to its biochemical composition.
5. The mass spectrum is compared to a library of reference spectra obtained from identical analyses of known microbes.
6. Once a match is found between the mass spectrum from the unknown and a reference mass spectrum, identification can be made.
-
- Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF)
- An automated approach to compare the mass spectrum of a specimen to a database of known mass spectra.
3. FAME Analysis
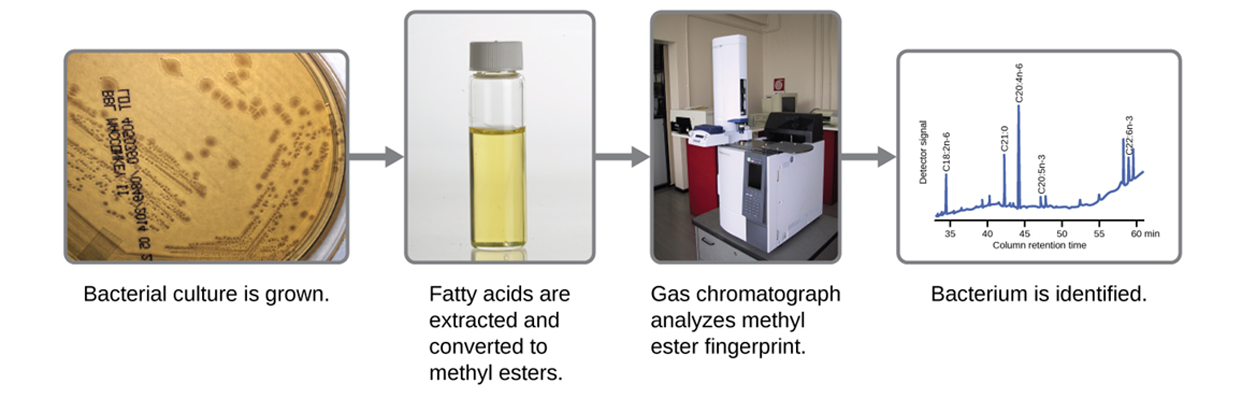
The MALDI-TOF analysis uses the mass spectrum of a microbe for identification, but other approaches use the unique lipid profiles of microbes to identify them. This requires analysis of the fatty acids present in the microbial membranes. One common approach used for this purpose is fatty acid methyl ester (FAME) analysis. The procedure used is described in the steps and shown in the image below.
-
1. Fatty acids are extracted from the membranes of microbes.
2. The extracted fatty acids are chemically altered to form volatile methyl esters.
3. Gas chromatography (GC), which is used to detect chemical components of a sample, is used to analyze the methyl esters.
4. The resulting GC chromatogram is compared with reference chromatograms.
5. Once a match is found between the GC chromatogram from the unknown and a reference chromatogram, the identification can be made.
Phospholipid-derived fatty acids (PFLA) analysis uses FAME analysis on fatty acids that have been extracted from membranes using saponification (hydrolysis with a strong base). This follows the same procedure described above after the fatty acids have been isolated for analysis.
-
- Fatty Acid Methyl Ester (FAME) Analysis
- An approach that analyzes the fatty acids present in microbial membranes and uses this for identification.
- Phospholipid-Derived Fatty Acids (PFLA) Analysis
- An approach that uses FAME analysis on fatty acids that have been extracted from membranes using saponification.
4. Proteomic Analysis
Proteomic analysis examines the proteins produced under specific growth conditions (the proteome). The steps of the analysis are summarized below.
-
1. Proteins are separated using high-pressure liquid chromatography (HPLC).
2. The collected fractions are digested to yield smaller peptide fragments.
3. Mass spectrometry is used to identify the peptide fragments.
4. The mass spectrum is compared with those of known microbes.
5. Once a match is found between the mass spectrum from the unknown and a reference mass spectrum, the identification can be made.
-
- Proteomic Analysis
- Examines the proteins produced under specific growth conditions (the proteome).
- Proteome
- Proteins produced under specific growth conditions.
In this lesson, you learned some examples of the many ways in which biochemical analyses can be used to identify microbes. After an introduction to the use of biochemistry for identification, you learned about general approaches such as detecting the presence or absence of compounds within the cell and examining microbial metabolism. Next, you learned the steps of MALDI-TOF analysis, FAME analysis, and proteomic analysis. The MALDI-TOF, FAME, and proteomic analyses use comparison of samples to reference data to identify microbes. These types of analyses are important because it can be difficult to quickly identify microbes, but identification is critical for diagnosis, disease treatment, and other purposes.


